Acute myeloid leukemia (AML) is the second most common form of childhood leukemia and has the worst prognosis of all major childhood cancers. Improving the treatment outcome for patients with AML remains a major clinical challenge. The nucleoside analog, cytarabine (Ara-C), has been the mainstay of AML chemotherapy for more than 40 years. However, extensive inter-patient variation in treatment response, development of resistance, and severe toxicity remain as major hurdles to effective Ara-C chemotherapy. It is a pro-drug that requires activation to Ara-CTP by multiple phosphorylation steps. Thus, inter-patient variation in gene expression levels of candidate genes involved in cytarabine metabolism can impact therapeutic outcome. In last few years' significant efforts have been invested in generating genomic, transcriptomic and epigenetic data in various cancer patients including AML. TCGA (The Cancer Genome Atlas) is one of the most commonly available such resource, recently another resource available to scientific community TARGET (Therapeutically Applicable Research to Generate Effective Treatments), a project initiated by the National Cancer Institute (NCI) in 2006. This databased was created with the aim to take multi-omics integrative approach to characterize molecular alterations associated with poor outcomes in some common children's cancers including AML. In this study, we leveraged TARGET database to extract gene expression levels (generated using Affymetrix Gene ST Array) to investigate whether inter-patient differences in expression levels of 11 key candidate genes in cytarabine metabolic pathway are predictive of minimal residual disease (MRD: positive or negative) and thus outcome. These genes included DCK and CMPK1 encoding for drug activating enzymes, CDA, DCTD, NT5C2, NT5C3A, important for cytarabine inactivation; drug influx transporter SLC29A1; RRM1 and RRM2 as ribonucleotide inhibitors; CTPS1 and NME3 which play a key role in the synthesis of nucleoside tri-phosphates. We also investigated correlation of the expression levels of these genes with each other to get insight into whether these genes could be co-regulated. In order to have consistency we only included data (gene expression and clinical and patient demographic, characteristics) from 105 AML patients where expression levels were available from bone marrow specimens and also restricted our analysis to AAML03P1 and AAML0531 clinical cohorts. Our resultsshow significant correlation in expression levels between cytarabine metabolic pathway genes resulting in three clusters: First cluster included DCK, NT5C2, NT5C3A, RRM1 and CMPK1 with positive correlation with each other's ranging between (r=0.619 to 0.720 p-value <0.0001). Second cluster included CDA, NME3 and SLC29A1 (r=0.186 to 0.438 p-value <0.05) and third cluster included CTPS1 and DCTD with positive correlation with each other.
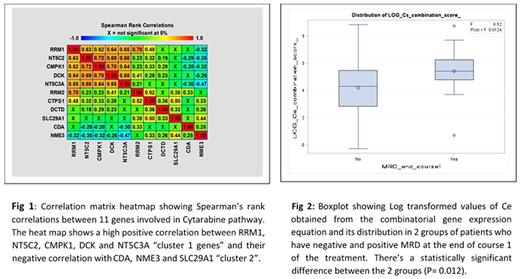
Our results from association analysis with MRD showed that cluster 1 genes had higher expression levels in patients with positive MRD. On the other hand, cluster 2 genes have all a higher expression in the MRD negative. To get the combinatorial effect of gene-expression on different clinical endpoints we defined a combination score (CS) using
CS = (DCK*NT5C2*NT5C3A*CMPK1*CTPS1*RRM1*RRM2*DCTD)/(CDA*NME3*SLC29A1).
Higher CS was associated with positive MRD (P= 0.012) (Figure 2) and lower overall survival rates (r= - 0.232, P-value = 0.017). By generating a combinatorial expression score for cytarabine pathway genes, we have been able to identify features that might predict MRD, overall survival and thus outcome in patients treated with cytarabine. An interesting fact that we observed positive correlations between activating genes - DCK, CMPK with inactivating genes such as NT5C2 or reductases RRM1/RRM2 that negatively impact the drug activation. One possible explanation, when DCK is highly expressed, CMPK1 responsible for the second phosphorylation step increases. At the same time, RRM1 and RRM2 known as DCK inhibitors and NT5C2 become highly expressed to balance high DCK level. This might also be a plausible reason for not observing consistent correlations between expression levels of ara-C pathway genes with outcome.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal